OpenAIRE Research Graph » History » Revision 30
« Previous |
Revision 30/36
(diff)
| Next »
Claudio Atzori, 10/11/2021 02:11 PM
The OpenAIRE Research Graph¶
- Table of contents
- The OpenAIRE Research Graph
The OpenAIRE Research Graph is one of the largest open scholarly record collections worldwide, key in fostering Open Science and establishing its practices in the daily research activities.
Conceived as a public and transparent good, populated out of data sources trusted by scientists, the Graph aims at bringing discovery, monitoring, and assessment of science back in the hands of the scientific community.
Imagine a vast collection of research products all linked together, contextualised and openly available. For the past ten years OpenAIRE has been working to gather this valuable record. It is a massive collection of metadata and links between scientific products such as articles, datasets, software, and other research products, entities like organisations, funders, funding streams, projects, communities, and data sources.
As of today, the OpenAIRE Research Graph aggregates around 450Mi metadata records with links collecting from 10K data sources trusted by scientists, including:- Repositories registered in OpenDOAR or re3data.org
- Open Access journals registered in DOAJ
- Crossref
- Unpaywall
- ORCID
- Microsoft Academic Graph
- Datacite
After cleaning, deduplication, enrichment and full-text mining processes, the graph is analysed to produce statistics for OpenAIRE MONITOR (https://monitor.openaire.eu), the Open Science Observatory (https://osobservatory.openaire.eu), made discoverable via OpenAIRE EXPLORE (https://explore.openaire.eu) and programmatically accessible as described at https://develop.openaire.eu.
Json dumps are also published on Zenodo.
TODO: image of high-level data model (entities and semantic relationships, we can draw here: https://docs.google.com/drawings/d/1c4s7Pk2r9NgV_KXkmX6mwCKBIQ-yK3_m6xsxB-3Km1s/edit)
Graph Data Dumps¶
In order to facilitate users, different dumps are available. All are available under the Zenodo community called OpenAIRE Research Graph.
Here we provide detailed documentation about the full dump:
- Json dump: https://doi.org/10.5281/zenodo.3516917
- Json schema: https://doi.org/10.5281/zenodo.4238938
Json schema¶
FAQ¶
Graph provision processes¶
TODO:3. Processes
- harvesting NOT SURE WHAT IOANNA WANTS: is what we have on graph.openaire.eu OK?
- transformation NOT SURE WHAT IOANNA WANTS: is what we have on graph.openaire.eu OK?
doiboost(not in processing, in #Aggregation business logic by major sources)- direct zenodo updates
- deduplication STARTED
- inference
- Funder ingestion ("Harry is speaking with funders, gets the list of projects, inference rules, ...")
For the processes we need a description of what it does, what's the input, which part of the graph it affects, and anything of importance ("if there is no input value for X, then we assume Y and assign the value Z to A")
OpenAIRE entity identifier and PID mapping policy
Aggregation business logic by major sources¶
TODO:2. input sources
- repositories
- journals
DOIBoost- *
MAG - *
Crossref - *
Unpaywall - projects
- organizations
- openorgs
- open citations
- openAPC
- etc etc
For each input source class, we need to know what's in there, the format, approximate numbers, peculiarities, important aspects of the data (e.g. "Crossref provides us with the authoritative list of publishers!"). Anything else of importance?
DOIBoost is the intersection among Crossref, Unpaywall, Microsoft Academic Graph and ORCID
DOIBoost¶
Datacite¶
EuropePMC¶
The strategy for the resolution of links between publications and datasets is defined by Scholexplorer
Deduplication business logic¶
Deduplication business logic for research results¶
Metadata records about the same scholarly work can be collected from different providers. Each metadata record can possibly carry different information because, for example, some providers are not aware of links to projects, keywords or other details. Another common case is when OpenAIRE collects one metadata record from a repository about a pre-print and another record from a journal about the published article. For the provision of statistics, OpenAIRE must identify those cases and “merge” the two metadata records, so that the scholarly work is counted only once in the statistics OpenAIRE produces.
Duplicates among research results are identified among results of the same type (publications, datasets, software, other research products). If two duplicate results are aggregated one as a dataset and one as a software, for example, they will never be compared and they will never be identified as duplicates.
OpenAIRE supports different deduplication strategies based on the type of results.
Methodology overview
The deduplication process can be divided into two different phases:- Candidate identification (clustering)
- Decision tree
- Creation of representative record
The implementation of each phase is different based on the type of results that are being processed.
Strategy for publications
Candidate identification (clustering)
Clustering is a common heuristics used to overcome the N x N complexity required to match all pairs of objects to identify the equivalent ones. The challenge is to identify a clustering function that maximizes the chance of comparing only records that may lead to a match, while minimizing the number of records that will not be matched while being equivalent. Since the equivalence function is to some level tolerant to minimal errors (e.g. switching of characters in the title, or minimal difference in letters), we need this function to be not too precise (e.g. a hash of the title), but also not too flexible (e.g. random ngrams of the title). On the other hand, reality tells us that in some cases equality of two records can only be determined by their PIDs (e.g. DOI) as the metadata properties are very different across different versions and no clustering function will ever bring them into the same cluster. To match these requirements OpenAIRE clustering for products works with two functions:- DOI: the function generates the DOI when this is provided as part of the record properties;
- Title-based function: the function generates a key that depends on (i) number of significant words in the title (normalized, stemming, etc.), (ii) module 10 of the number of characters of such words, and (iii) a string obtained as an alternation of the function prefix(3) and suffix(3) (and vice versa) o the first 3 words (2 words if the title only has 2). For example, the title “Entity deduplication in big data graphs for scholarly communication” becomes “entity deduplication big data graphs scholarly communication” with two keys key “7.1entionbig” and “7.1itydedbig” (where 1 is module 10 of 54 characters of the normalized title.
Decision tree
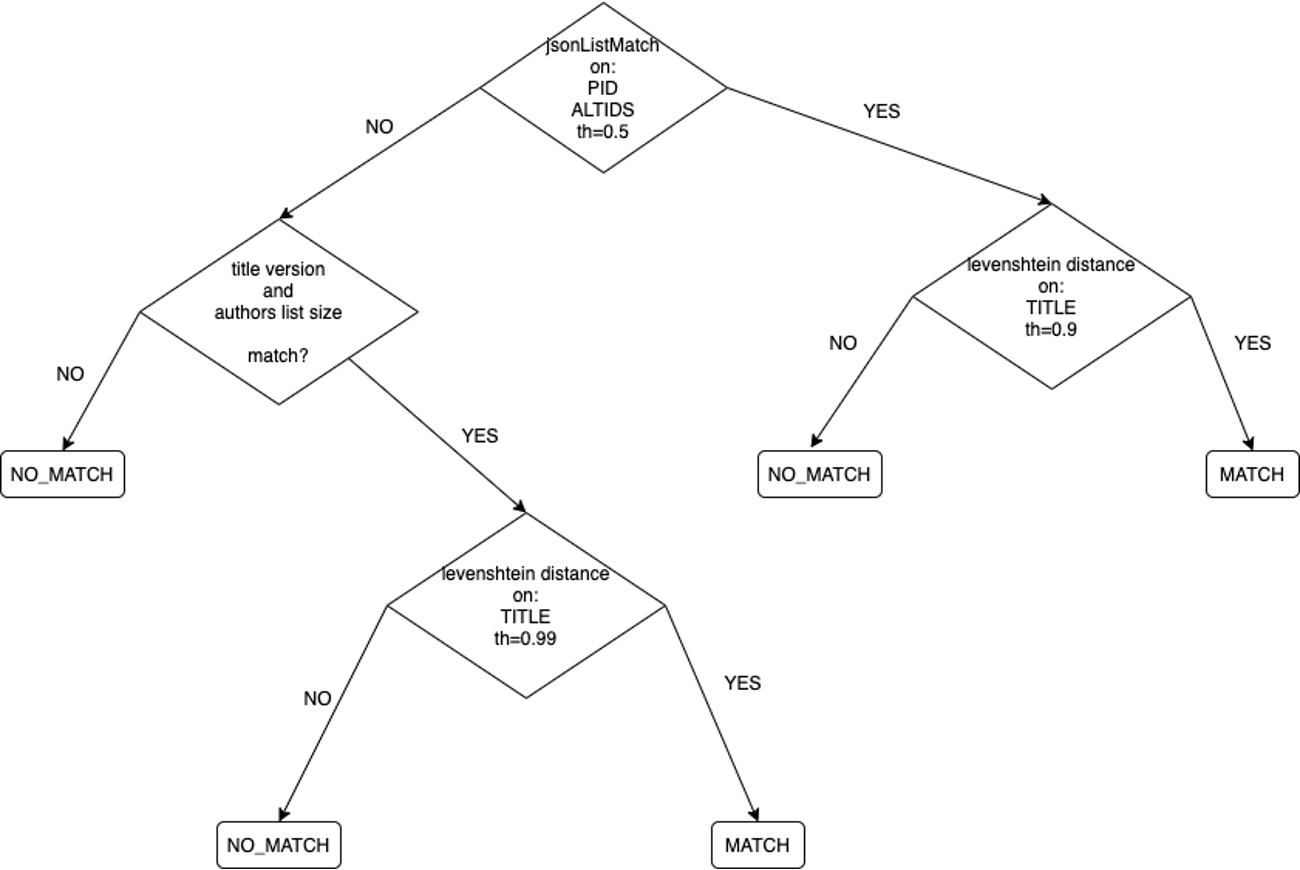
For each pair of publications in a cluster the following strategy (depicted in the figure below) is applied.
Cross comparison of the pid lists (in the pid and alternateid elements). If 50% common pids, levenshtein distance on titles with low threshold (0.9).
Otherwise, check if the number of authors and the title version is equal. If so, levenshtein distance on titles with higher threshold (0.99).
The publications are matched as duplicate if the distance is higher than the threshold, in every other case they are considered as distinct publications.
Creation of representative record
TODO
Strategy for datasets
Strategy for software
Strategy for other types of research products
Clustering functions
NgramPairs
It produces a list of concatenations of a pair of ngrams generated from different words.
Example:
Input string: “Search for the Standard Model Higgs Boson”
Parameters: ngram length = 3
List of ngrams: “sea”, “sta”, “mod”, “hig”
Ngram pairs: “seasta”, “stamod”, “modhig”
SuffixPrefix
It produces ngrams pairs in a particular way: it concatenates the suffix of a string with the prefix of the next in the input string.
Example:
Input string: “Search for the Standard Model Higgs Boson”
Parameters: suffix and prefix length = 3
Output list: “ardmod” (suffix of the word “Standard” + prefix of the word “Model”), “rchsta” (suffix of the word “Search” + prefix of the word “Standard”)
TODOs¶
- OpenAIRE entity identifier & PID mapping policy (started, to be completed by Claudio and/or Michele DB)
- Aggregation business logic by major sources:
Unpaywall integrationCrossref integrationORCID integrationCross cleaning actions: hostedBy patch- Scholexplorer business logic (relationship resolution)
- DataCite
- EuropePMC
- more….
- Deduplication business logic (started, to be completed by Michele DB)
- For research outputs (
publications, datasets, software, orp) - For research organizations
- For research outputs (
- Enrichment
- Mining business logic
- Deduction-based inference
- Propagation business logic
- Post-cleaning business logic
- FAQ
Updated by Claudio Atzori over 4 years ago · 30 revisions